So we’ve established that it’s often difficult to get an estimate of species’ environmental tolerances experimentally. What’s the alternative? Well, one alternative is to look at the set of conditions under which our species occurs naturally. When scientists go out to collect or observe organisms in the wild, they frequently provide specimens or observational data to museums or other data storage centers. Part of the data that is often submitted is locality data – where did you see/catch the species in question? This data used to be in the form of verbal descriptions or rough estimates of latitude and longitude from maps, but nowadays is more often provided as GPS coordinates. It turns out that this data is very useful in estimating species environmental tolerances and preferences.
In the most general sense, here’s how niche modeling works. First, we have a set of occurrence points for our species:
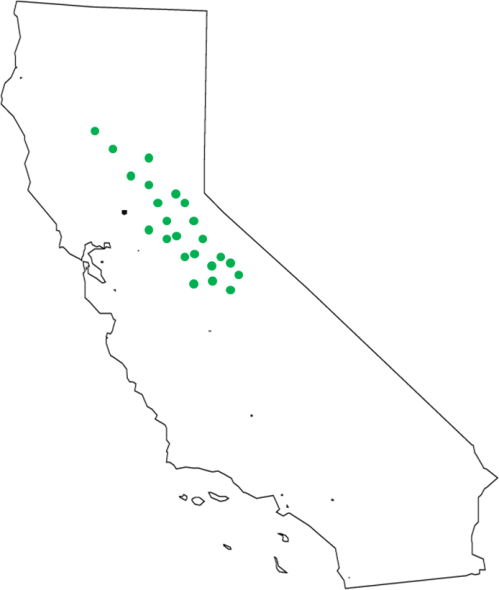
Each of these occurrences represents an observation of a set of environmental conditions under which we know our species can persist. Unfortunately, though, this occurrence data almost never comes with direct estimates of all of the environmental factors which are potentially relevant to determining the distribution of the species. However, we now have very fine-scale data sets containing localized estimates of a bunch of different environmental factors over the entire planet. We can use that data in conjunction with our occurrence data to estimate the sets of conditions under which our species has been found, by extracting the environmental conditions present at each of those occurrence points.
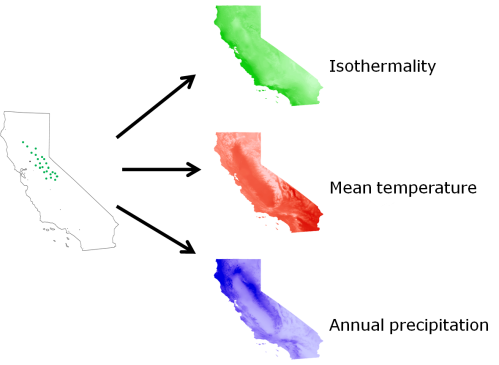
So where we once just had a bunch of latitude and longitude measurements, we now have a set of conditions that we know our species can handle, at least in the short term (if you look back at the definition of the fundamental niche you’ll see a problem here, but we’ll get to that later). This is a valuable resource, because we can use these estimates to start building mathematical models of the environmental tolerances of our species. There are a zillion different methods for building these models, but that’s a discussion for another time. Suffice to say that we now have a set of points in environment space that we have extracted from our points in geographic space. Like so:

Environmental niche modeling is simply the application of some algorithm to estimate species tolerances from this sort of data. They range from very simple heuristics, such as “take the central 95% interval of the species distribution for each environmental variable”, which looks something like this:

To highly complex methods based on algorithms from machine learning that compare the points where the species can be found to the sets of habitats in which they have not been found:

Once we’ve got our model, we can project it back onto the geographic distribution of environmental variables to estimate the suitability of habitat across the entire geographic space. In summary, the process looks something like this:

Where the colors on the final figure represent the estimated suitability of habitat for our species (note that in this case I just picked a model from the pile I had for California, it is not based on the points in the illustration).
This is a tremendously powerful thing, to the extent that our models can be trusted – it not only tells us something about the environmental tolerances of our species, it can tell us where we might look for populations that we haven’t seen yet. We can also take the mathematical model of the species tolerances that we constructed in this process and project it onto environmental conditions in other geographic regions to model the ability of the species to invade other areas, or even project it onto estimated conditions in the past or future to estimate the historical or future distributions of species given various scenarios of climate change.
I have glossed over some very serious methodological and conceptual issues with niche modeling in this post, but this is intended as an introduction only. I’ll get to the ugly bits later. There are also some serious methodological issues that arise with transferring models to other geographic regions or time periods, but we’ll save those for another day too. For now we’ll concentrate on the contrast between these methods and the experimental physiological methods I discussed in my last Serious Science Post (i.e., not the post about iguanas farting in the bathtub or various creatures delivering interspecific high fives).
Remember that one of the key issues with physiological niche estimates is their tractability – it is very difficult to get fine-scale estimates of the limits of species tolerances, and becomes increasingly difficult with the addition of more variables. That’s not anywhere near as much of an issue here – all we have to do to get a new data point for our species is to see it while we have our GPS in hand. That’s a wonderful thing, because it means that we can amass large amounts of data with minimal effort. Since environmental variables vary over space, we can also get estimates of the response of our species to a great number of environmental variables at once with the same set of occurrence data.
So what’s the catch? Well, it actually turns out that there are a heck of a lot of them. I’ll probably do several posts on key methodological issues with niche models, but I’ll start just by pointing this one out: not all combinations of environmental variables occur in the real world. Going back to our diagram from before:

We see that our species is perfectly happy in the hottest extremes of our environment space (i.e., the farthest to the right). What if they were happy in even hotter temperatures? We have no way of knowing whether that’s true or not, because we’ve already found them in the hottest places available to us! Likewise, maybe they can handle higher precipitation levels in those hotter climates than they can in colder ones – it certainly looks like there might be a positive correlation between their tolerances for precipitation and temperature. Once again, we have no way of knowing because there are no extremely hot and extremely wet regions from which we could possibly sample (or fail to sample) our species. If we were taking an experimental approach we could simply expand the range of conditions we were testing our species under, but that doesn’t work here – as my mother always told me, we can’t afford to heat the whole outdoors.
This is a serious issue – it means that our ability to estimate the niche is limited not only by our number of occurrence points, but by the distribution of environmental variables in the study region. This issue is serious enough that it has led some people to suggest that we not think of these methods as niche estimates at all, referring to them instead as “species distribution models”. While I’m sympathetic to that, I still use the term “niche model” for reasons I’ll clarify in a later post.
In summary, niche modeling overcomes a lot of the difficulties that arise with physiological estimation of the niche, but brings a whole slew of other issues along with it. TANSTAAFL, as always.







